BALSA / ELSA
BALSA / ELSA is an economic, fast and accurate solution for the secondary analysis of next generation sequencing data.
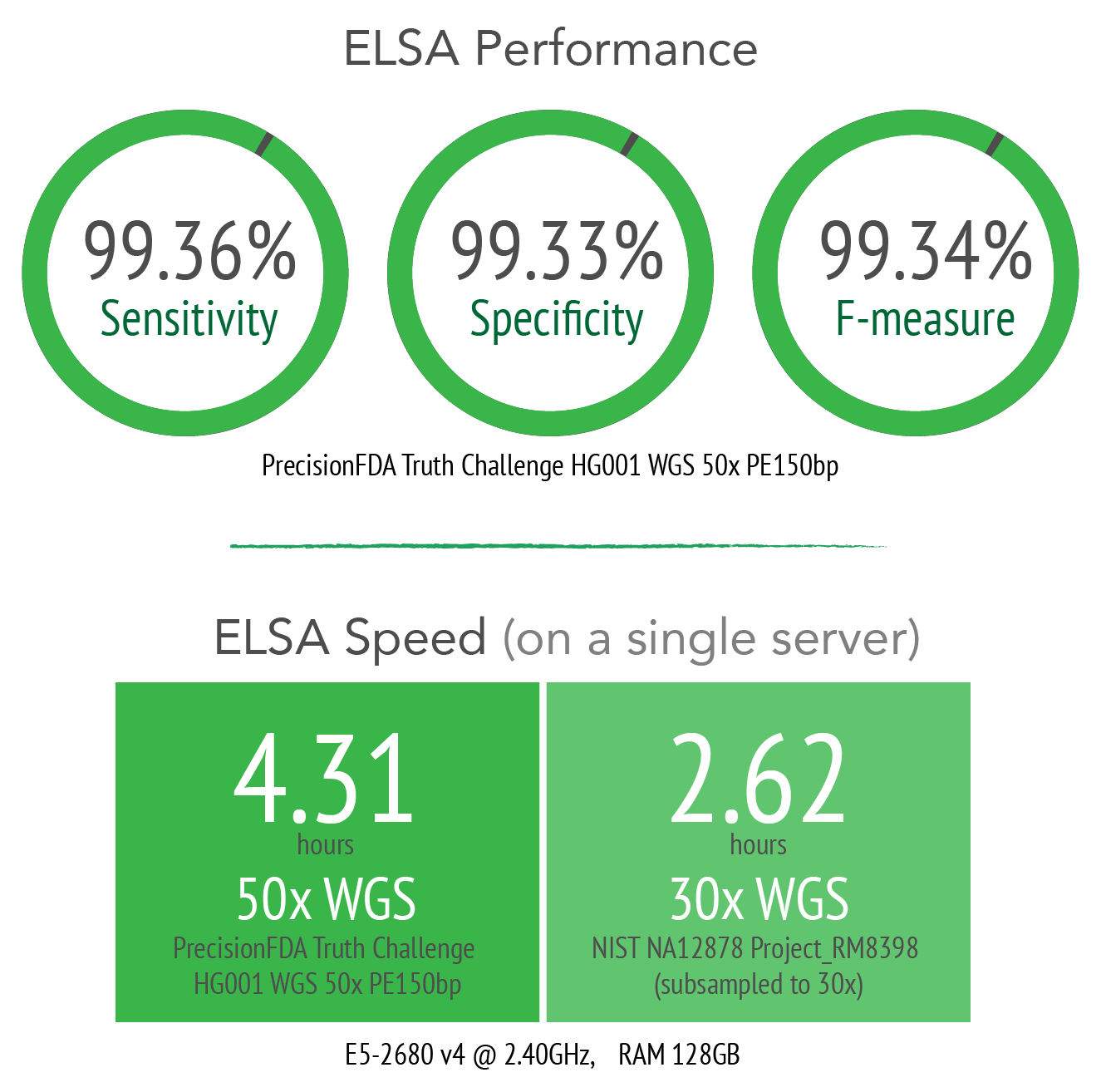
BALSA / ELSA* can process a 50-fold whole genome sequence (WGS) sample from raw reads to variants, including single nucleotide polymorphism (SNP), and insertion and deletion (INDEL), in about 4 hours, or just 10 minutes for a 200-fold whole exome sequence (WES) sample.
- WGS alignment and variant calling
- WES alignment and variant calling
- Amplicon sequencing
- Tumor pairs
- Variant outputs in VCF format
- Alignment outputs in BAM or SNAPSHOT formats
* BALSA requires GPU support while ELSA is a version of BALSA optimized to run on multi-core CPUs only.
BALSA / ELSA takes advantage of highly optimized indexing and parallel algorithms to effectively exploit a GPU and CPU SIMD, respectively, to speed up processes like alignment, score recalibration, INDEL realignment, de-duplication and variant calling.
A 16-genotype model is incorporated to support the calling of SNPs and INDELs. Validation of the final results with the NIST Genome In A Bottle standard demonstrates that BALSA / ELSA's analysis pipeline has the highest combined sensitivity and precision.

BALSA can run at a minimum hardware requirement of 6- core CPU and an NVIDIA GPU with 4GB RAM, while ELSA needs a minimum 16-core CPU with 64GB RAM.
See the table below to find out which version is more suitable for your current hardware.
Different kinds of licensing are available to suit your infrastructure. See the table below.
| ELSA | BALSA-Power8 | BALSA-x86 | |
| Minimum Hardware Requirement |
8-core CPU 64GB RAM |
Power8 S814L 64GB RAM NVIDIA GPU with 4GB RAM |
6-core CPU 64GB RAM NVIDIA GPU with 4GB RAM |
| Recommended Hardware Requirement | 16-core CPU 96 GB RAM |
Power8 S824L 96 GB RAM NVIDIA GPU with 6GB RAM |
12-core CPU 96 GB RAM NVIDIA GPU with 6GB RAM |
BALSA / ELSA Benchmark Results on GIAB
* Luo R, Wong Y, Law W, Lee L, Cheung J, Liu C, Lam T. (2014) BALSA: integrated secondary analysis for whole-genome and whole-exome sequencing, accelerated by GPU.PeerJ 2:e421 dx.doi.org/10.7717/peerj.421