
Building Secure, Scalable Platforms for Genomics and Clinical Data
See our WorksSee ProductsOur team are actively publishing and contributing to cutting-edge AI applications in healthcare.
Clair is a powerful variant calling tool that uses deep learning to detect SNPs and indels from sequencing data, especially from long-read technologies like Oxford Nanopore and PacBio. Designed for speed and accuracy, Clair handles the high error rates of sequencing data with ease, making it ideal for research and clinical applications. Its neural network model learns directly from sequencing data, enabling precise variant detection even in complex genomic regions. Fast, scalable, and open-source, Clair is a go-to solution for modern genomics workflows. Whether for large-scale studies or individual analyses, Clair delivers reliable results with minimal setup.




Our interdisciplinary team at the University of Hong Kong excels in integrating advanced machine learning with clinical expertise to tackle complex healthcare challenges. In the study, we employed reinforcement learning to optimize long-term cardiovascular disease prevention strategies, demonstrating our capability to harness AI for impactful health solutions. By leveraging extensive health data and cutting-edge analytics, we provide innovative, data-driven approaches to improve patient outcomes. Our commitment to excellence in research and technology positions us at the forefront of digital health innovation.
Our team at the University of Hong Kong pioneers the integration of large language models (LLMs) with clinical genomics to streamline variant interpretation. Through the development of AutoPM3, we automate the extraction of ACMG/AMP PM3 evidence from scientific literature, combining optimized retrieval-augmented generation (RAG) systems and TableLLM with Text2SQL capabilities. Evaluated on the PM3-Bench dataset, AutoPM3 significantly outperforms existing methods in variant identification. By wrapping AutoPM3 with a user-friendly interface, we enhance its accessibility, demonstrating our commitment to advancing AI-driven precision medicine

We built a secure online workspace that lets research teams safely access sensitive health and genetic data, while fully protecting patient privacy. The platform allows easy setup of research environments with all the tools scientists need, right from their browser. Data can be brought in or taken out through a secure review process, and access is carefully managed so only the right people can see the right information. With built-in safety checks, approval workflows, and detailed tracking, this system helps organizations stay compliant while enabling smooth, trustworthy collaboration in cutting-edge health research.
We built an advanced platform that helps professionals make sense of complex genetic data—especially when searching for clues behind rare diseases, cancer, or inherited conditions. Designed for both clinical and research use, it supports real-time manual review of over 10 million genetic variants per case, delivering results in under a minute in full production environments. The platform brings together data from trusted medical and research sources, making it easier to spot meaningful changes in DNA. It supports both individual and large-scale group studies, helping uncover hidden patterns and rare findings across families or populations. With powerful search, smart filters, and interactive visual tools, users can dig deep into the data and generate clear, actionable reports. Throughout the process, patient privacy and data security are fully protected. Whether you're studying drug responses or discovering new gene-disease links, this platform makes complex analysis faster, easier, and more insightful.


Our data analysis platform powers the secure, high-throughput processing of genomic and clinical data. Built on the Helicube platform, the Platform enables encrypted FASTQ uploads via a web interface with auto-resume and real-time status updates. Clinical and lab data are retrieved through integrated APIs. Behind the scenes, a robust 50-server cluster processes over 5TB of genetic data daily, ensuring rapid turnaround for high-volume next-generation sequencing (NGS) analysis. The platform stores results in enterprise-grade hot storage for real-time access, while archiving to AWS Glacier (HK) for high-redundancy backup. It orchestrates complex NGS workflows—like BWA+GATK and supports custom pipeline development. With smart recovery, user activity logging, and a visual pipeline editor, the Platform offers flexibility and resilience at scale. Whether running standard or custom analyses, the Platform delivers performance, transparency, and reliability—empowering precision genomics with confidence.
We developed Clinical Data Portal to give healthcare professionals a clear and secure view of each patient’s medical journey. Through one centralized platform, Clinical Data Portal brings together essential health records—including doctor notes, lab samples, medication history, and family background—making it easier for teams to deliver personalized care. Clinical Data Portal also supports key workflows like patient registration and withdrawal, reducing manual steps and minimizing errors. By integrating with other systems, Clinical Data Portal ensures that all relevant data—clinical, genetic, and laboratory—is connected and available when it’s needed most. With strong privacy and security protections built in, Clinical Data Portal helps clinicians work efficiently and confidently, while ensuring patients’ information stays safe. It’s a smarter, more connected way to support modern, data-driven healthcare.
To support our advanced healthcare and genomics applications, we’ve built and maintained a powerful data infrastructure spanning over 5 petabytes—that’s five million gigabytes—across multiple secure locations. This robust data farm ensures that critical clinical and genomic data is always available when it’s needed most. With over 99.99% uptime, our systems run reliably around the clock, supporting high-speed analysis, secure storage, and smooth collaboration across healthcare teams. This infrastructure powers the largest genomic institute in Hong Kong, enabling researchers and clinicians to work with massive volumes of data—quickly, safely, and without interruption. Whether it's processing sequencing data, managing patient records, or supporting real-time decision-making, our data systems are designed for performance, scalability, and resilience. By investing in enterprise-grade hardware and cloud technologies, we ensure that our platform not only keeps up with today’s demands, but is also ready for the future of precision medicine.
MegaHit is a NGS de novo assembler for assembling large and complex metagenomics data in a time- and cost-efficient manner. It finished assembling a soil metagenomics dataset with 252 Gbps in 44.1 and 99.6 h on a single computing node with and without a graphics processing unit, respectively. MegaHit assembles the data as a whole, i.e. no pre-processing like partitioning and normalization was needed. When compared with previous methods on assembling the soil data, MegaHit generated a three-time larger assembly, with longer contig N50 and average contig length; furthermore, 55.8% of the reads were aligned to the assembly, giving a fourfold improvement.
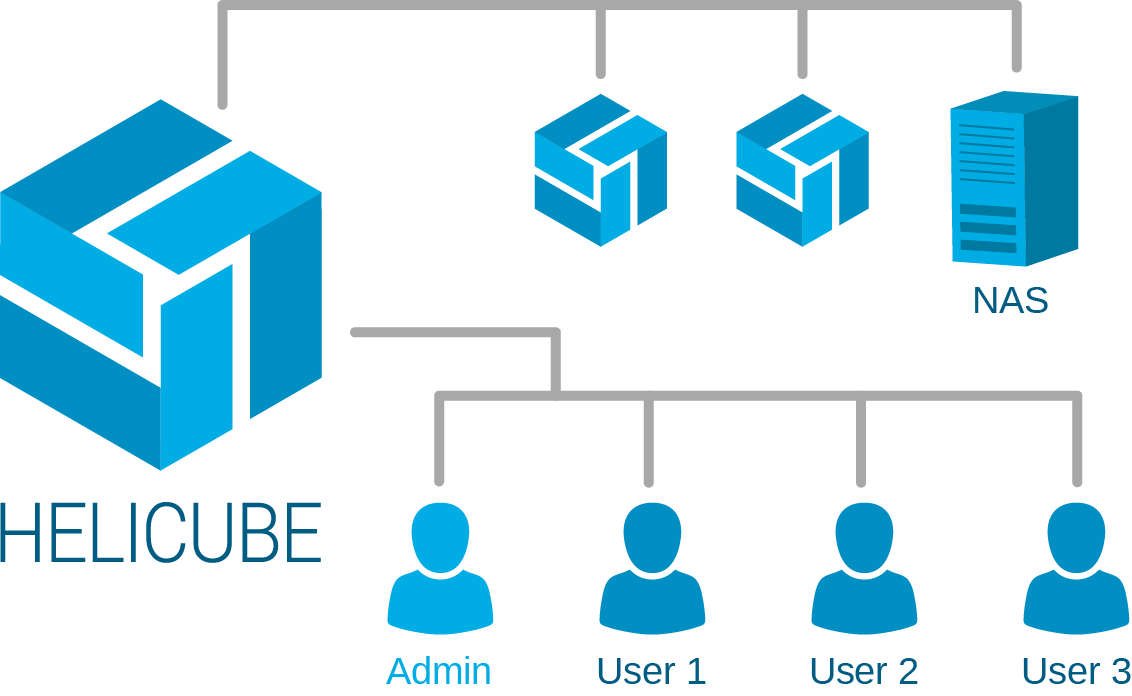
Helicube schedules and spreads out jobs to the worker units in the cluster. It then executes the jobs in parallel to achieve horizontal scalability.
Helicube can adapt new analysis tools and configure them to run on its platform.
Monitor system health, loading speed and utilisation using Helicube's administration panel.
database.bio delivers genomic insights into genetics research and everyday medical practice through flexible, robust and secure web application.
database.bio uses a configurable decision tree to help rapidly prioritise genetic variants into five pathogenicities.
Each genetic variant is presented with human-readable annotations with links to external supporting evidence.
Includes over 30 databases and 120 annotation types for analysing panel, exome, or genome variants.
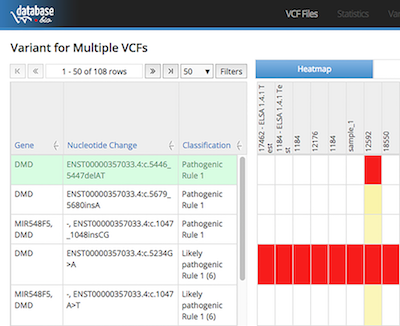
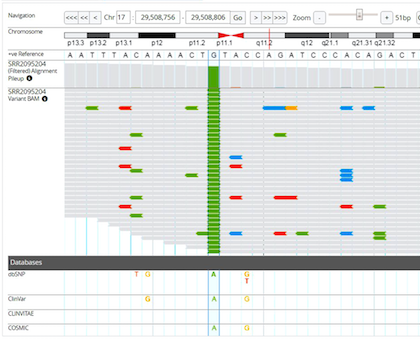
MGB (Multi Genome Browser) is a web application that visualizes different analysis result such as variants, BAM alignments, alignments pileup, BED, primer and the reference. It provides insight into alignments quality and the variant characteristics. It also supports feature of multiple tracks to show several BAM files, pileups and sets of BED regions. MGB can be set up in a files hub and serveral workstations can link to them without duplicating the result files. With fast response and short loading time, MGB is a good support application for any bioinformatics institutes, clinics and hospitals.
BALSA/ELSA allows organisations to analyse data quickly without additional investments in large computing systems.
Takes only 4.08 hours to process a 50-fold whole genome sequence (WGS) sample, or 10 minutes for a 200-fold whole exome sequence (WES) sample.
Validation with the NIST Genome In A Bottle standard demonstrates that BALSA/ELSA has the highest combined sensitivity and specificity.
Outputs variants in VCF, alignment results in SAM/BAM format and stores a SNAPSHOT for efficient storage and indexing.
BGI Online helps you to do genome sequencing and analysis projects, while eliminating software setup and file transfer logistics.
It is a secure cloud platform for bioinformaticians to advance life-saving research.
Access the world’s largest repository of genetic data.
Create your own pipelines or use our best practices.
Share your findings to clients and peers.
Visualise your results using database.bio.

