database.bio
database.bio is an application that combines variant annotation, categorization, and visualization so as to provide insight into individual genetic characteristics. It integrates five important domains related to clinical diagnosis (namely, variants, genes, diseases, drugs, and pathways) and assigns severity levels to variants using customizable categorization rules. Variant information is presented in HTML pages that contain annotation details using a powerful embedded genome browser (MGB), allowing clinical practitioners and biological researchers to carry out analysis and comparison of genomic data in a highly configurable manner.
Biomedical databases:
database.bio supports the integration of more than 30 biomedical databases for the purpose of variant annotation, which includes variant functional prediction, variant conservation, splicing prediction, regulatory feature prediction, real sample data such as 1000G, TCGA, ICGC, and LOVD, as well as clinical trial details.
Designed for medical practitioners:
With a simple setup, database.bio provides straight-through processing of sequencer’s raw data into presentable analysis results. Users are free from tedious and time-consuming manual work like web searches, database digging, and information aggregation. database.bio also supports comparative studies of multiple samples, e.g. patients versus controls, patients versus their parents or relatives.
Customizable rule-based categorization:
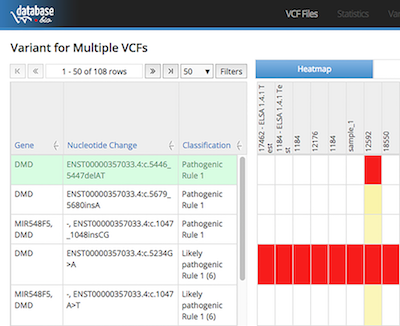
To assist medical practitioners to sort out the important variants from the many others, database.bio categorizes variants into 5 severity levels (Pathogenic, Likely-pathogenic, VOUS, Likely-benign, and Benign), based on categorization rules that can be customized by medical practitioners to exploit their insight and practice in integrating the information from various biomedical databases.
An ultra-fast genome viewer:
MGB, database.bio’s embedded genome viewer, can smoothly display the alignment of reads for multiple samples (specifically, up to 10,000 reads per sample, while using only 500MB of memory on the client machine). MGB provides different views of variants based on multiple samples and biomedical databases. It runs on web browsers with cross-platform support and no installation required.